Eight plots (selectable by which) are currently available using ggplot or graphics:
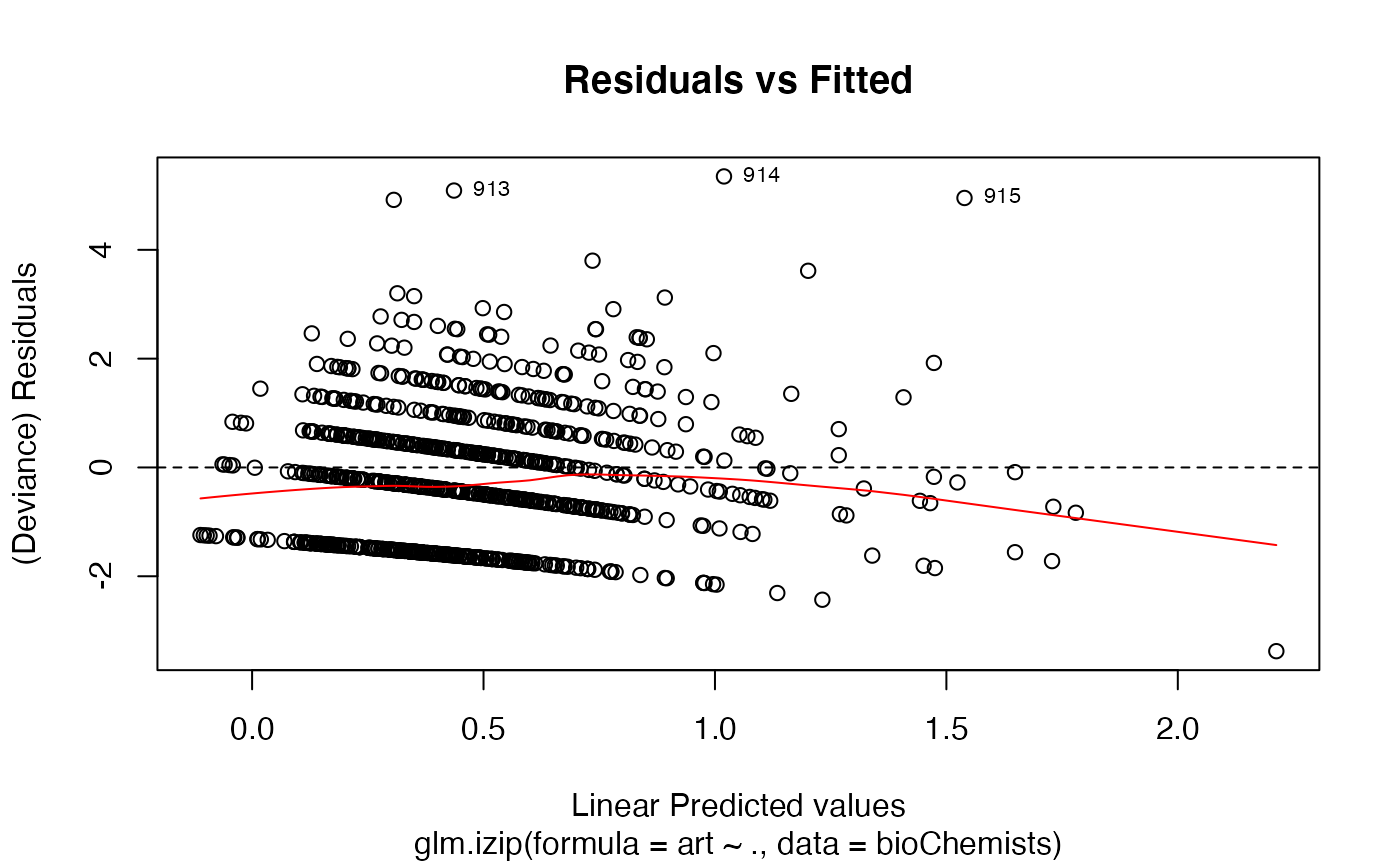
a plot of deviance residuals against fitted values,
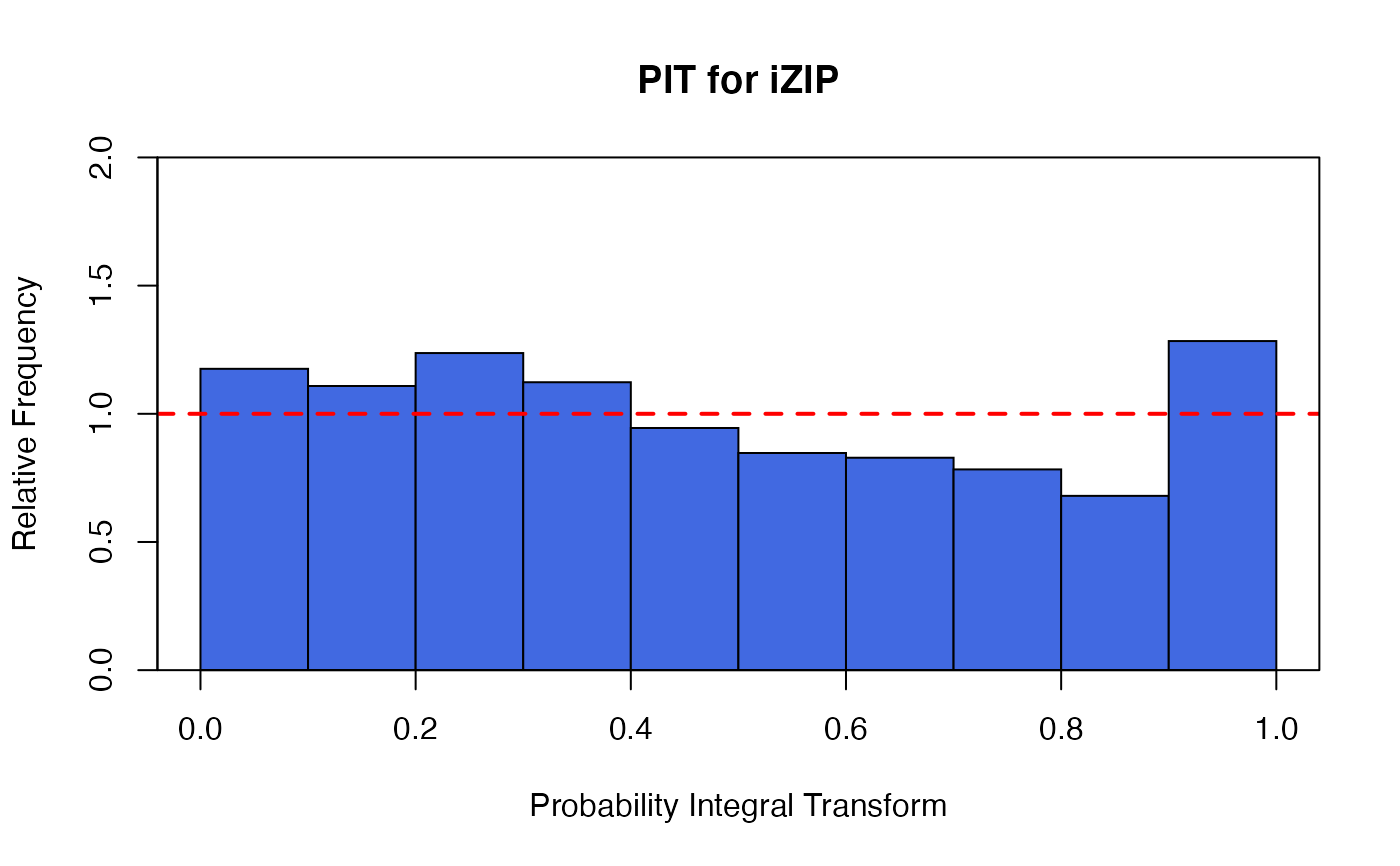
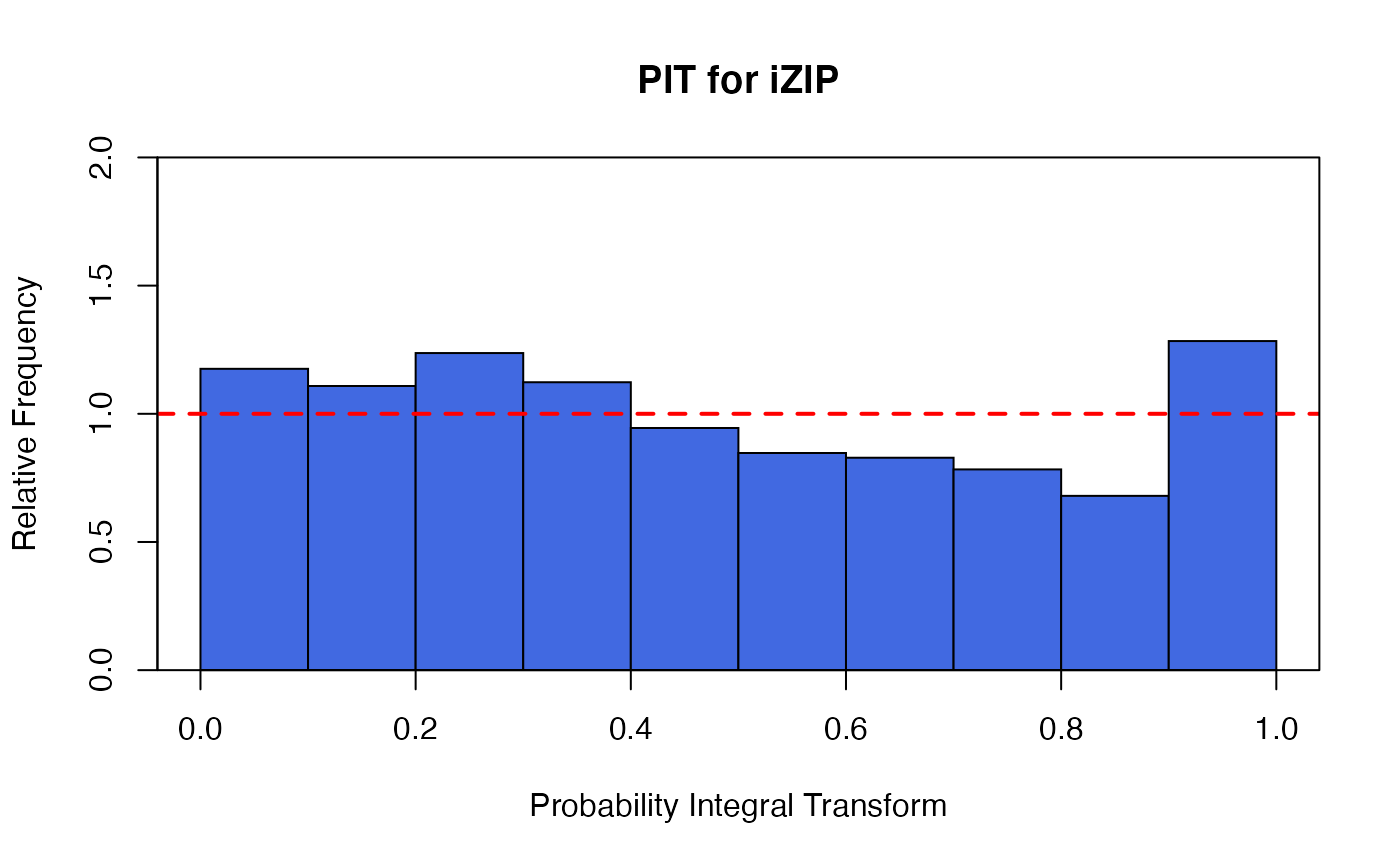
a non-randomized PIT histogram,
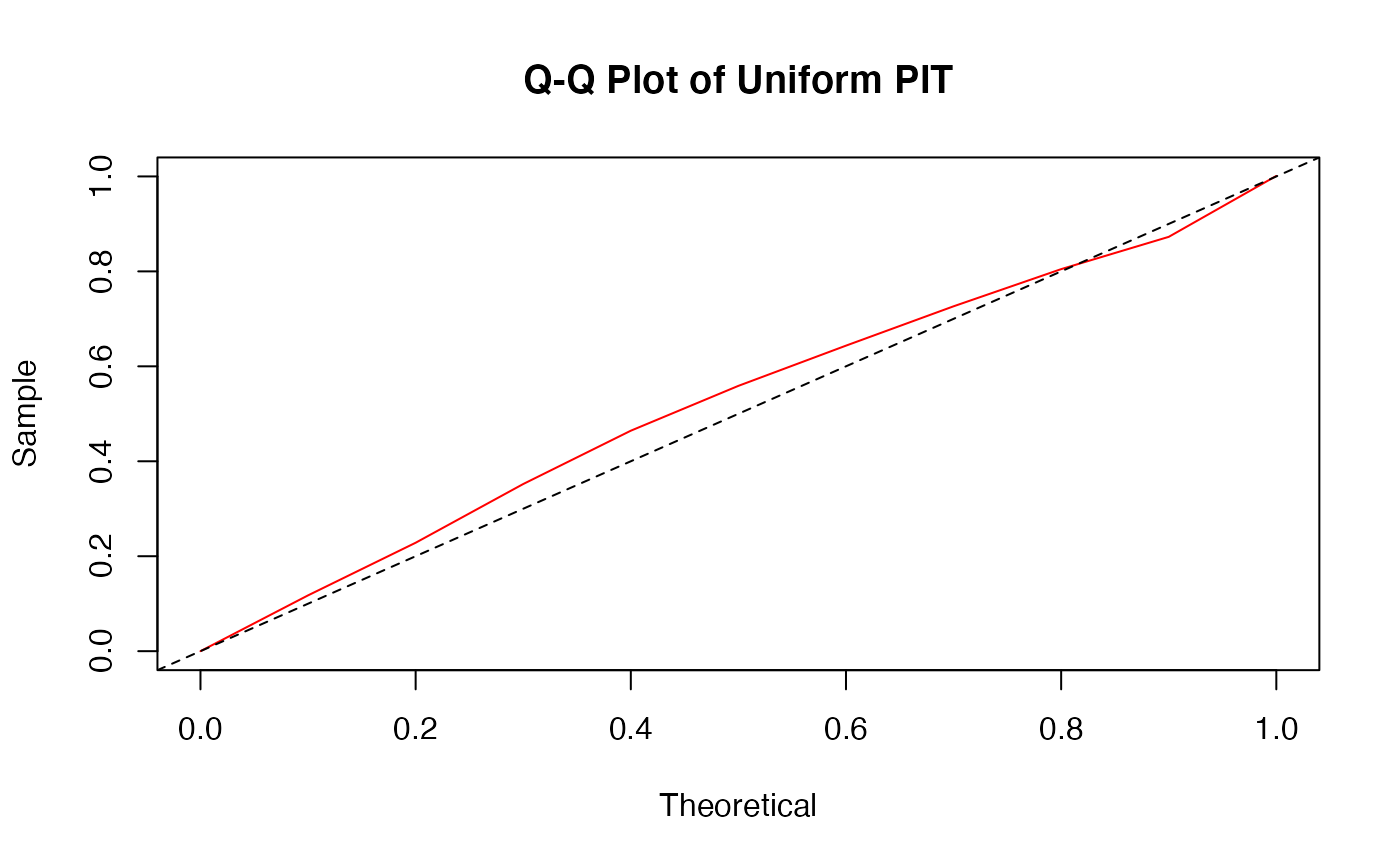
a uniform Q-Q plot for non-randomized PIT,
a histogram of the normal randomized residuals,
a Q-Q plot of the normal randomized residuals,
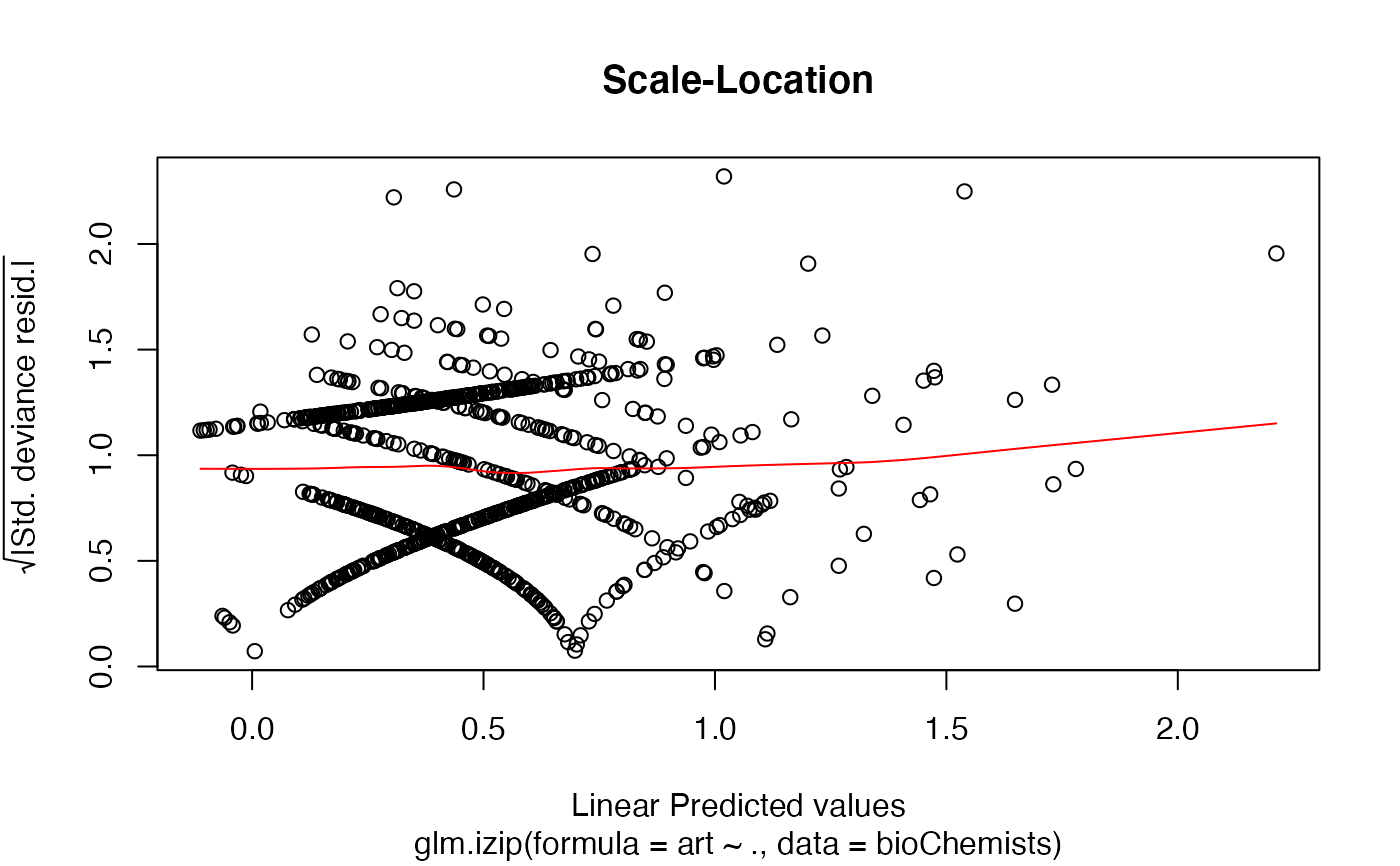
a Scale-Location plot of sqrt(| residuals |) against fitted values
a plot of Cook's distances versus row labels
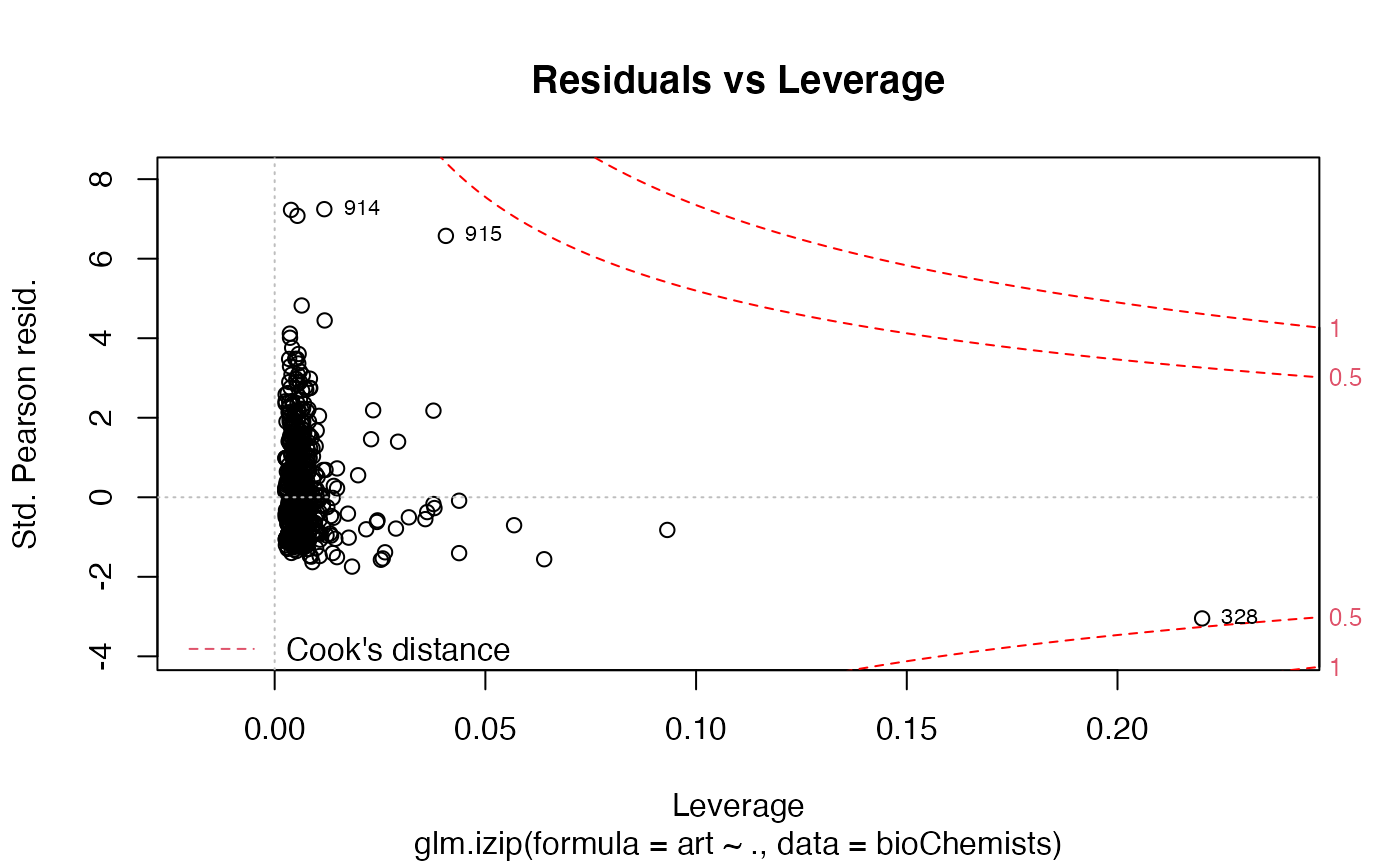
a plot of pearson residuals against leverage.
By default, four plots (number 1, 2, 6, and 8 from this list of plots) are provided.
# S3 method for izip plot( x, which = c(1L, 2L, 6L, 8L), ask = prod(par("mfcol")) < length(which) && dev.interactive(), bins = 10, ... ) # S3 method for izip autoplot( x, which = c(1L, 2L, 6L, 8L), bins = 10, ask = TRUE, nrow = NULL, ncol = NULL, output_as_ggplot = TRUE )
Arguments
| x | an object class 'izip' object, obtained from a call to |
|---|---|
| which | if a subset of plots is required, specify a subset of the numbers 1:8. See 'Details' below. |
| ask | logical; if |
| bins | numeric; the number of bins shown in the PIT histogram or the PIT Q-Q plot. |
| ... | other arguments passed to or from other methods (currently unused). |
| nrow | numeric; (optional) number of rows in the plot grid. |
| ncol | numeric; (optional) number of columns in the plot grid. |
| output_as_ggplot | logical; if |
Value
A ggarrange object, which is a ggplot or a list of ggplot for autoplot.
Details
The 'Scale-Location' plot, also called 'Spread-Location' plot, takes the square root of the absolute standardized deviance residuals (sqrt|E|) in order to diminish skewness is much less skewed than than |E| for Gaussian zero-mean E.
The 'Scale-Location' plot uses the standardized deviance residuals while the Residual-Leverage plot uses the standardized pearson residuals. They are given as \(R_i/\sqrt{1-h_{ii}}\) where \(h_{ii}\) are the diagonal entries of the hat matrix.
The Residuals-Leverage plot shows contours of equal Cook's distance for values of 0.5 and 1.
There are two plots based on the non-randomized probability integral transformation (PIT)
using izipPIT. These are a histogram and a uniform Q-Q plot. If the
model assumption is appropriate, these plots should reflect a sample obtained
from a uniform distribution.
There are also two plots based on the normal randomized residuals calculated
using izipnormRandPIT. These are a histogram and a normal Q-Q plot. If the model
assumption is appropriate, these plots should reflect a sample obtained from a normal
distribution.
See also
izipPIT, izipnormRandPIT,
and glm.izip.
Examples
data(bioChemists) M_bioChem <- glm.izip(art ~ ., data = bioChemists) ## The default plots are shown plot(M_bioChem) # or autoplot(M_bioChem)# or autoplot(M_bioChem, which = c(2,3))